Metabolomics Workflow (v2.1.0)¶
Summary¶
The gas chromatography-mass spectrometry (GC-MS) based metabolomics workflow (metaMS) has been developed by leveraging PNNL’s CoreMS software framework. The current software design allows for the orchestration of the metabolite characterization pipeline, i.e., signal noise reduction, m/z based Chromatogram Peak Deconvolution, abundance threshold calculation, peak picking, spectral similarity calculation and molecular search, similarity score calculation, and confidence filtering, all in a single step.
Workflow Diagram¶
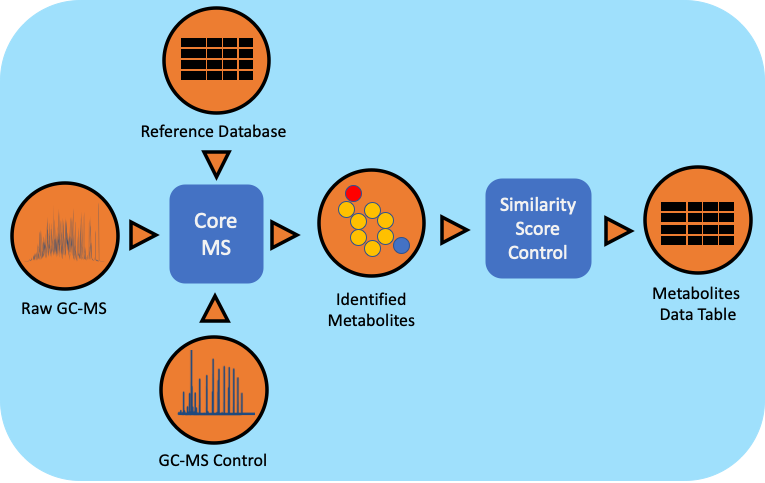
Workflow Dependencies¶
Third party software¶
- CoreMS (2-clause BSD)
- Click (BSD 3-Clause “New” or “Revised” License)
Database¶
- PNNL GC-MS Spectral Database
Workflow Availability¶
The workflow is available in GitHub: https://github.com/microbiomedata/metaMS
The container is available at Docker Hub (microbiomedata/metaMS): https://hub.docker.com/r/microbiomedata/metams
The python package is available on PyPi: https://pypi.org/project/metaMS/
The databases are available by request. Please contact NMDC (support@microbiomedata.org) for access.
Test datasets¶
https://github.com/microbiomedata/metaMS/blob/master/data/GCMS_FAMES_01_GCMS-01_20191023.cdf
Execution Details¶
Please refer to:
https://github.com/microbiomedata/metaMS/blob/master/README.md#usage
Inputs¶
- ANDI NetCDF for GC-MS (.cdf)
- CoreMS Hierarchical Data Format (.hdf5)
- CoreMS Parameter File (.JSON)
- MetaMS Parameter File (.JSON)
Outputs¶
- Metabolites data-table
- CSV, TAB-SEPARATED TXT
- HDF: CoreMS HDF5 format
- XLSX : Microsoft Excel
- Workflow Metadata:
- JSON
Requirements for Execution¶
- Docker Container Runtime
- Python Environment >= 3.6
- Python Dependencies are listed on requirements.txt
Version History¶
- 2.1.0
Point of contact¶
Package maintainer: Yuri E. Corilo <corilo@pnnl.gov>